The End-It™ DNA End-Repair Kit is used to convert DNA containing damaged or incompatible 5'- and/or 3'-protruding ends that result from shearing, restriction endonuclease digestion or PCR amplification to 5'-phosphorylated, blunt-end DNA for fast and efficient blunt-end cloning into plasmid, cosmid, fosmid, BAC or other vectors. The conversion to blunt-end DNA is accomplished by exploiting the 5'→3' polymerase and 5'Æ3' exonuclease activities of T4 DNA Polymerase. T4 Polynucleotide Kinase and ATP are also included in the kit for phosphorylation of the 5'-ends of the blunt-ended DNA for subsequent ligation into a cloning vector. The End-It™ DNA End-Repair Kit is identical to the End-Repair Enzyme Mix provided in the pWEB™ Cosmid Cloning Kit, the pWEB-TNC™ Cosmid Cloning Kit, and the EpiFOS™ Fosmid Library Production Kit and the CopyControl™ Fosmid Library Production Kit.
The End-It™ DNA End-Repair Kit contains reagents for 20 end-repair reactions (repair of up to 100µg of genomic DNA). The end-repaired, 5'-phosphorylated DNA can be efficiently ligated into a blunt-end, dephosphorylated cloning vector using the Fast-Link™ DNA Ligation Kit.
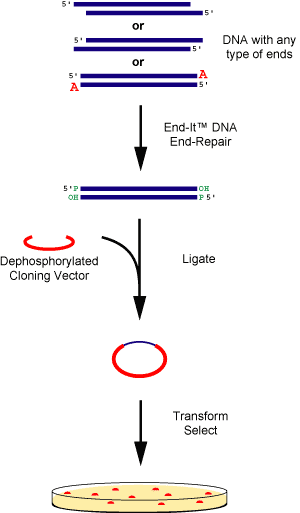 |
Figure 1. DNA fragments containing any type of ends are rapidly and efficiently converted to 5'-phosphorylated, blunt-ended DNA using the End-It™ DNA End-Repair Kit. The end-repaired DNA is ready for blunt-end ligation into the cloning vector of choice using, for example EPICENTRE®'s Fast-Link™ DNA Ligation Kit. |
If you cannot find the answer to your problem then please contact us or telephone +44 (0)1954 210 200