Ribonuclease R (RNase R) from E. coli is a magnesium-dependent 3´→5´ exoribonuclease that digests essentially all linear RNAs but does not digest lariat or circular RNA structures1,2, or doublestranded RNA with 3´ overhangs shorter than seven nucleotides.2 Most cellular RNAs will be digested completely by RNase R, with the exception of tRNAs, 5S RNA, and intron lariats (Fig. 1). The 3´ tails of lariats will be trimmed by RNase R to the branch point nucleotide, where there is a 2´,5´-phosphodiester linkage. Lariats are produced during pre-mRNA splicing of intron regions and can be isolated from a mixture of total RNA by digestion with RNase R. The ArrayPure™ Kit, and MasterPure™ RNA and Yeast RNA Purification Kits are ideal for such total RNA preparations.
RNA isolated using this method can be used as a template to produce labelled cDNA as a target for microarrays containing potential intron sequences, or for tiling arrays containing overlapping regions of complete chromosomes or genomes. The cDNA produced will not be a linear representation of the intron, but the sequences contained in it will be intron-derived.
Unit Definition
One unit of RNase R converts 1 µg of poly(A) into acid-soluble nucleotides in 10 minutes at 37°C under standard assay conditions.
Storage Buffer
RNase R is supplied in a 50% glycerol solution containing 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 0.1 mM EDTA, 1 mM DTT, and 0.1% Triton® X-100.
RNase R 10X Reaction Buffer
0.2 M Tris-HCl (pH 8.0), 1 M KCl, and 1 mM MgCl2.
Note
RNase R requires low (0.1-1.0 mM) magnesium concentrations for activity. Low EDTA concentrations in substrate RNA solutions can negatively affect RNase R activity. Additional MgCl2 up to 1 mM final concentration can be used to compensate for EDTA in the substrate. Optimal activity is at 37°C.
Quality Control
RNase R is function-tested in a reaction containing a mixture of linear and circularized RNA oligonucleotides. Only the linear RNA is digested.
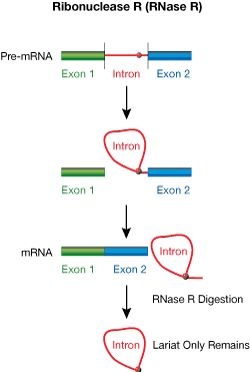 |
| Figure 1. Schematic overview showing processing of intron lariats by RNase R |
If you cannot find the answer to your problem then please contact us or telephone +44 (0)1954 210 200