
MagIC Beads
The technology behind Element Zero targeted RNA and DNA capture
Magnetic Instant Capture Beads (MagIC Beads) based systems offer unmatched efficiency in hybridization-based, sequence-specific capture of nucleic acids for various research applications.
MagIC Beads are an attractive alternative to classical approaches that utilize a combination of biotinylated oligonucleotide probes and streptavidin-coated beads for the enrichment of nucleic acids.
One technology, multiple applications:
RNA-protein interactome
Mass-spectrometry
Western blotting
Silver staining on a protein gel
RNA Enrichment
RNA-RNA interactions
RNA modifications
Alternative polydenylation
Alternative splicing
Alternative transcription start sites
Native elongating transcript sequencing (NET-seq)
DNA Enrichment
DNA epigenetic modifications
Chromosomal rearrangements
Genome integration sites (viruses, transposons, genetic engineering etc.)
Detection of mutations
The Beads
DNA probes covalently attached to the beads:
The beads are produced through a novel approach of direct synthesis of DNA capture probes on the surface of functionalized magnetic nanoparticles. The synthesis reaction produces a high number of oligonucleotide chains, which are covalently attached to the surface of the particles through their 5’ ends.
Arrays of probes with different sequences on a single population of the particles:
The process allows for the simultaneous, unbiased synthesis of complex arrays of capture probes with different sequences on a single population of the particles. All of the probes from the array are present on the surface of each particle in a 1:1 ratio.
Bind to multiple regions of a target:
The beads developed for the capture of a specific sequence carry an array of different probe sequences which target multiple regions of the same target DNA or RNA molecule. The fact that those unique probes are clustered together in close proximity to one another allows them to act in synergy in binding the target (hybridization of a single probe brings the target within close proximity to other capture probes). This effect leads to rapid and very stable binding of the beads to the target molecules and provides a marked advantage over approaches using arrays of biotinylated capture probes in which no probe synergy is observed.
No steric hindrance effect due to long molecular linker:
The proprietary functional modification of the particles also provides an exceptionally long molecular linker, which provides an appropriate distance between the surface of the particles and the sequence of the capture probes. This property eliminates the steric hindrance effect, which is known to have the potential of interfering with the ability of the capture probe to correctly hybridize with its complementary sequence.
Graphical representation of the MagIC Bead design:
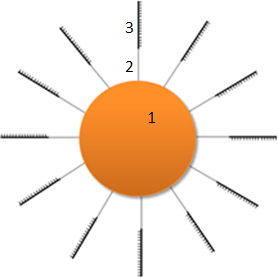
1 – Magnetic particle.
2 – Long molecular linker.
3 – Capture probe sequences. The capture probes attached to the bead surface can represent any number of unique sequences, which are evenly spread and represented on the beads at equal ratios to one another.
The Probes
We design the probes for you:
MagIC Beads based systems are supplied with the capture probes designed by ElementZero Biolabs relieving the researchers from the burden of having to design their own capture probes. The probe design strategy has been optimized for the unique properties of the system and ensures full compatibility with the bead and buffer design provided by the company. ElementZero Biolabs offers its wealth of experience to provide arrays of efficient capture probes screened and selected against potential off-target binding in any organism with annotated genome and/or transcriptome for any sequence of interest.
High specificity, precise capture – 25-40nt probes:
The probes present in each array provided by ElementZero have varying lengths (typically 25-40nt) but have a strict, standardized GC content, hybridization temperature, and specificity towards the intended target. All of those properties have been extensively standardized for ensuring optimal performance in various experimental setups.
The Buffers
Optimized for each application:
MagIC Beads based kits are provided with buffers specifically optimized for various applications, ensuring optimal efficiency and specificity in different aims and environments of the capture. In classic workflows utilizing biotinylated oligonucleotides in combination with streptavidin beads, the environment of the capture reaction has to allow for the stable binding of biotin to streptavidin. This strictly limits the possibility of applying the conditions which would be particularly beneficial for the capture from the perspective of probe hybridization.
The best conditions for the capture of unfragmented nucleic acids:
Traditional workflows:
Salt concentration impairs the hybridization of the target sequences to capture probes:
Unlike for MagIC beads, in traditional workflows salt concentration has to be high enough to preserve the stability of biotin-streptavidin interactions in elevated temperatures. Buffers with high salt concentration are used (500 mM or more), leading to the stabilization of secondary structures formed by potential nucleic acid targets, in turn preventing the hybridization of the target sequences to the capture probes, rendering the system unreliable for the capture of highly structured or long, unfragmented transcripts and DNA segments.
Low concentration of chaotropic agents increases unspecific probe binding:
Chaotropic agents can disrupt biotin-streptavidin interactions. High concentrations of chaotropic agents, however, have beneficial properties for the regulation of nucleic acids base pairing. Certain agents can regulate the temperature in which probes will hybridize to the complementary sequences and help disrupt the hybridization between sequences with imperfect complementarity, resulting in a reduction of the binding of the probes to off-target sequences.
Low concentration of denaturing agents that preserve the integrity of target nucleic acid:
The high concentration of denaturing agents in the buffers in combination with other buffer components can ensure deactivation of nucleases, thus ensuring the integrity of assayed nucleic acids throughout the process of the target capture. Denaturing agents also help ensure full disruption of protein-nucleic acid interactions, which are undesirable in some experimental setups.
But with MagIC Beads:
Low salt concentration – capture unfragmented nucleic acids:
Employ buffers with relatively low salt concentration, which in combination with other buffer components allow for reliable capture of unfragmented nucleic acid targets independently of their length or secondary structure.
Optimized concentrations of chaotropic agents:
Buffers supplied with MagIC Beads contain well-optimized concentrations of chaotropic agents, utilizing their properties to provide superb probe hybridization specificity and efficiency.
Leverage the benefits of denaturing factors:
MagIC Buffers contain optimized concentrations of denaturing factors. As a result, MagIC Beads based kits do not require the use of nuclease inhibitors in capture reactions DNase and RNase inhibition is provided by the composition of the buffers) and allow for the capture of target nucleic acids free from other types of molecules.
Contact Cambio for a custom quote for 16, 24, 48 or 96 reactions today:
support@cambio.co.uk / Tel: 01954 210200
