ELASA: Aptamer-Based ELISA
Wednesday, 26 June 2019
 Enzyme-Linked Immuno-Sorbent Assays (ELISA) were first developed in 1971, shortly before the development of the hybridoma method for producing monoclonal antibodies. Since that time, antibody-based ELISAs have been widely used for specific, sensitive detection of key biomarkers or molecules of interest. ELISA involves binding of an antigen (molecule or target of interest), or antibody then antigen, to a plate. This is followed by binding of a labeled antibody for detection. Sandwich assays typically utilize two antibodies binding to different epitopes on the antigen for capture and detection. There are several methods that can be used to immobilize antibodies or aptamers on a plate and a wide range of dyes and reagents that can be employed for detection (3).
Enzyme-Linked Immuno-Sorbent Assays (ELISA) were first developed in 1971, shortly before the development of the hybridoma method for producing monoclonal antibodies. Since that time, antibody-based ELISAs have been widely used for specific, sensitive detection of key biomarkers or molecules of interest. ELISA involves binding of an antigen (molecule or target of interest), or antibody then antigen, to a plate. This is followed by binding of a labeled antibody for detection. Sandwich assays typically utilize two antibodies binding to different epitopes on the antigen for capture and detection. There are several methods that can be used to immobilize antibodies or aptamers on a plate and a wide range of dyes and reagents that can be employed for detection (3).
Aptamers can be used in place of a capture antibody, detecting antibody, or both. Enzyme-Linked Aptamer-Sorbent Assays (ELASA) offer several key advantages. Because aptamers are chemically synthesized they can be produced quickly with excellent batch-to-batch consistency. They are easily modified, conjugated, and complexed with other molecules without loss of affinity or selectivity. Unlike traditional antibodies, aptamers can be selected for binding to non-immunogenic targets and selected to discriminate between highly simlar compounds. (3). The use of aptamers also eliminates non-specific binding caused by heterophilic antibodies, anti-animal antibodies, or rheumatoid factor (antibodies binding to the Fc region of IgG antibodies) that is frequently observed in antibody-based sandwich assays. (1,2).
Competitive ELASA for Detection of Canine CD40L
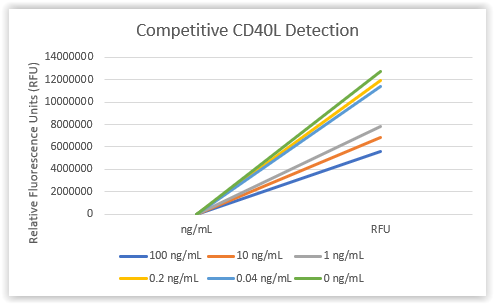
In a competitive assay, antigen is bound to the plate. Antigen in the sample competes with immobilized antigen for binding to a detecting antibody or aptamer. Increased concentration of antigen in the sample reduces the amount of detecting antibody or aptamer available to bind immobilized target and causes a reduction in signal. As part of an NIH SBIR contract, Base Pair is discovering affinity reagents to important canine cancer biomarkers and immuno-oncology targets. In preliminary testing, Base Pair has demonstrated detection of canine CD40L using a DNA aptamer in a competitive assay. Biotinylated canine CD40L aptamer was pre-incubated with varying concentrations of CD40L protein, then evaluated for binding to a CD40L-coated plate. Reduction in signal was observed for concentrations of 1 ng/mL, 10 ng/mL, and 100 ng/mL of CD40L protein in an initial prototype assay, with a minimum detectable concentration falling between 200 and 1,000 pg/mL. Aptamer concentration, buffer formulations, and detection reagents can be further optimized to enhance assay sensitivity.
Sandwich ELASA for Detection of Ebolavirus
Base Pair has been collaborating with Dr. Misaki Wayengera of Makerere University in Uganda to explore aptamers for the development of simpler, more sensitive assays for the diagnosis of Ebolavirus*.
Ebolavirus is a filovirus that causes severe hemorrhagic fever with a high rate of mortality. Believed to originate with the fruit bat, the virus can be transferred from infected animals and insects and also through the transfer of blood, mucus, or interaction with infected materials. On August 1, 2018, the Ministry of Health of the Democratic Republic of the Congo in Africa declared a new outbreak of Ebola Virus Disease (EVD) in the North Kivu Province. As of June 9, 2019, there have been 1,968 confirmed cases and 1,296 confirmed deaths(4). An experimental vaccine is being administered and a clinical trial for a new therapeutic is underway(5).
Base Pair has developed a prototype sandwich assay for the detection of Zaire Ebolavirus GP1 glycoprotein, found on the surface of the virion. The assay is based on the use of selective DNA aptamers for the capture and detection of Zaire Ebolavirus GP1 protein. Dilutions of culture supernatant containing inactivated Zaire Ebolavirus virion (supplied by John Klena, Ph.D. of the CDC) were successfully detected in the assay.
View Preliminary Data from Zaire Ebolavirus GP1 ELASA
*The current work and Dr. Wayengera's filovirus laboratory at Makerere University are funded by the European & Developing Countries Clinical Trials Partnership ( EDCTP). Grand Challenges Canada provided the initial seed funding for the pan filovirus RDT innovation in 2013.
References
1. Emerson, J.F. et al. Endogenous antibody interferences in immunoassays. Lab Medicine. 2013. 44(1):69-73.
2. Schwickart, M., et al. Interference in immunoassays to support therapeutic antibody development in preclinical and clinical studies. Bioanalysis. 2014. 6(14):1939-1951
3. Toh, S.ZY., et al. 2014. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosensors and Bioelectronics. 64:392-403.
4. WHO, Ebola situation reports: Democratic Republic of the Congo, accessed June 11, 2019, https://www.who.int/ebola/situation-reports/drc-2018/en/
5. WHO, Ebola virus disease fact sheet, accessed June 11, 2019, https://www.who.int/news-room/fact-sheets/detail/ebola-virus-disease
Article Source:
Base Pair Bio