Catalog Number: 10-1986-xx
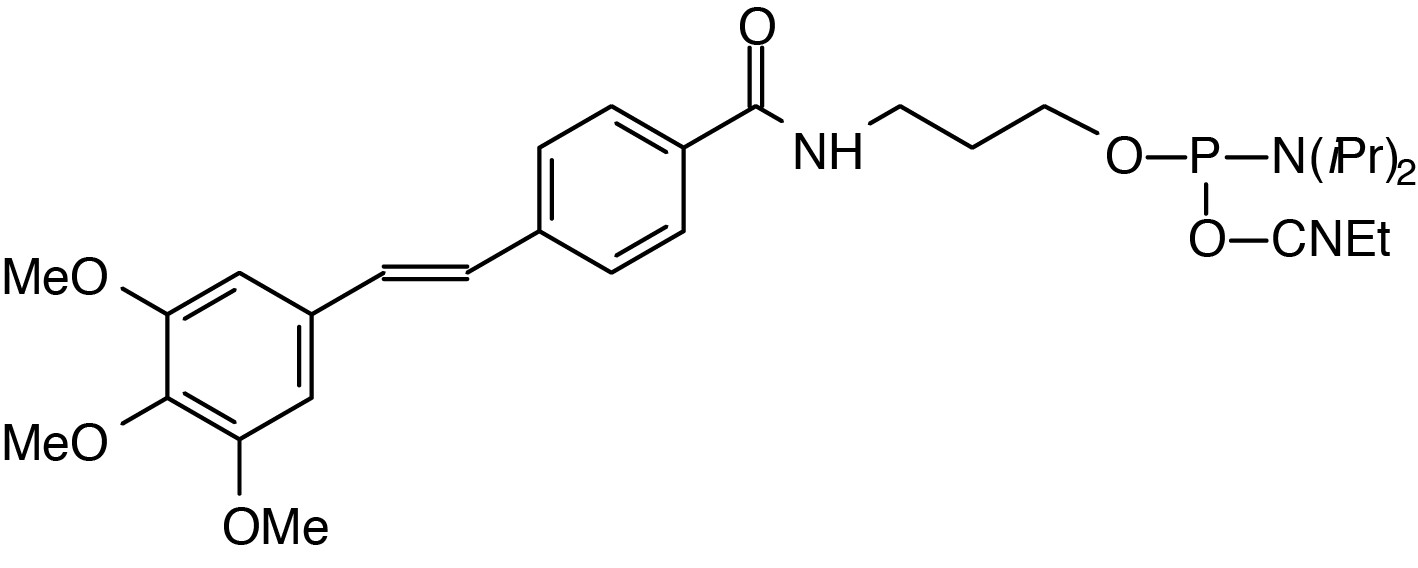
Description: 5'-Trimethoxystilbene Cap Phosphoramidite
(3,4,5-trimethoxystilbene-4'-yl)-(3-carboxamidopropyl)-1-O-(2-cyanoethyl)-
(N,N-diisopropyl)-phosphoramidite |
| Formula: C30H42N3O6P |
M.W.: 571.65 |
F.W.: 433.39 |
Diluent: Anhydrous Acetonitrile
Add fresh diluent to product vial to recommended concentration and swirl vial occasionally over several minutes until product is completely dissolved. (Some oils may require between 5 and 10 minutes.) Use care to maintain anhydrous conditions. In case of transfer to alternate vial type, ensure recipient vial has been pre-dried. For more information, see http://www.glenres.com/Technical/TB_ABITransfer.pdf.
|
| Coupling: No changes needed from standard method recommended by synthesizer. |
| Deprotection: No changes needed from standard methold recommended by synthesizer. |
| Storage: Freezer storage, -10 to -30°C, dry |
| Stability in Solution: 6 months |
Melting point increases of over 10 °C per modification can be realized for short duplexes.1,2 The caps fit canonical Watson-Crick base pairs and do not stack well on mismatched base pairs. This leads to increased base pairing selectivity at the terminal and the penultimate position of oligonucleotides featuring the caps. Base pairing fidelity is usually low at the termini, where fraying occurs frequently in the absence of caps. The beneficial effects of the caps are also realized when longer target strands are bound, so there is no need for blunt ends for the duplexes formed.1,2 The caps, when attached to the 5’ terminus of an oligonucleotide, also facilitate purification as their lipophilicity leads to prolonged retention on reversed phase columns or cartridges. Finally, capping of termini may discourage the degradation of oligonucleotides by exonucleases.
3’-Uaq Cap CPG, a Uridine support modified with a 2’- anthraquinone residue, is the most effective oligonucleotide cap known to date.3,4 For short hybrid duplexes between DNA probes and RNA target strands, the increase in Tm is up to 18 °C and the modification is effective in increasing the Tm of DNA:DNA, RNA:RNA, and DNA:RNA hybrid duplexes. 3’-Uaq Cap also increases probe specificity by depressing the melting point of terminal mismatches.
Caps for Increased Duplex Stability and Base-Pairing Fidelity at Termini
New cap structures allow for the preparation of hybridization probes with increased affinity for complementary sequences. The monomers used to prepare capped oligonucleotides are phosphoramidites that can be readily introduced via automated DNA synthesis at the end of solid phase syntheses. The caps favor the formation of stable Watson-Crick duplexes by stacking on the terminal base pair.
Melting point increases of over 10 °C per modification can be realized for short duplexes.1,2 The caps fit canonical Watson-Crick base pairs and do not stack well on mismatched base pairs. This leads to increased base pairing selectivity at the terminal and the penultimate position of oligonucleotides featuring the caps. Base pairing fidelity is usually low at the termini, where fraying occurs frequently in the absence of caps. The beneficial effects of the caps are also realized when longer target strands are bound, so there is no need for blunt ends for the duplexes formed.1,2 The caps, when attached to the 5’ terminus of an oligonucleotide, also facilitate purification as their lipophilicity leads to prolonged retention on reversed phase columns or cartridges. Finally, capping of termini may discourage the degradation of oligonucleotides by exonucleases.
3’-Uaq Cap CPG, a Uridine support modified with a 2’- anthraquinone residue, is the most effective oligonucleotide cap known to date.3,4 For short hybrid duplexes between DNA probes and RNA target strands, the increase in Tm is up to 18 °C and the modification is effective in increasing the Tm of DNA:DNA, RNA:RNA, and DNA:RNA hybrid duplexes. 3’-Uaq Cap also increases probe specificity by depressing the melting point of terminal mismatches.
If you cannot find the answer to your problem then please contact us or telephone +44 (0)1954 210 200