Ready-Lyse™ Lysozyme Solution is a non-mammalian, non-avian, recombinant lysozyme preparation for the lysis of Gram-negative (including E. coli) and Gram-positive (such as Bacillus sp.) bacteria. The use of Ready-Lyse™ Lysozyme results in higher yields of protein than obtained with standard egg white lysozyme, therefore, less is needed in a reaction. Also, unlike egg white lysozyme, Ready-Lyse Lysozyme Solution is stable at -20°C, eliminating the need to prepare a fresh solution for each use.

Table 1. Bacteria lysed with Ready-Lyse Lysozyme
| Gram-negative | Gram-positiv |
|---|
| Escherichia coli |
Oerskovia xanthinolytica |
| Salmonella typhimurium |
Bacillus subtillus |
| Actinobacillus pleuropneumoniae |
|
| Rhodobacter sphaeroides |
|
| Shewanella putrefaciens |
|
| Flavobacteria odoratum |
|
Using Ready-Lyse™ Lysozyme Solution for Nucleic Acid Extraction
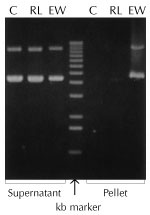 |
Figure 1. Lysis with Ready-Lyse™ Lysozyme increases yields of nucleic acids. 500µg per ml of pHC79 cosmid DNA was incubated for 15 minutes at 22°C in TE buffer (25mM Tris (pH 8.0), 10mM EDTA) containing either 5 µg (30 KU) per ml of Ready-Lyse Lysozyme or 500 µg (25 KU) per ml of egg white lysozyme. The solutions were centrifuged for 10 minutes and the pellets were resuspended in TE buffer containing 0.1% SDS. DNA in supernatants and pellets was separated by electrophoresis in a 0.8% agarose gel. Approximately 50% of the DNA was lost due to precipitation by egg white lysozyme (EW), while Ready-Lyse Lysozyme (RL) caused minimal precipitation losses of DNA compared to control (C) samples without lysozyme. |
Using Ready-Lyse™ Lysozyme Solution for Protein Extraction
 |
Figure 2. Use of Ready-Lyse™ Lysozyme Solution to recover recombinant proteins. One ml of induced cells from a recombinant E. coli clone was pelleted by microcentrifugation before induction and at 1 and 3 hours after induction. Each sample was resuspended in 50µl of cold TEBG buffer. One µl of Ready-Lyse Solution was added to each suspension and the cells were incubated at room temperature for 30 minutes. The cell debris was pelleted and 10 µl of the supernatant were run on an SDS-PAGE gel. Lane1, molecular weight markers; Lanes 2-4, time points of induction. The induced protein is designated by an arrow. |
Unit Definition
One unit of Ready-Lyse Lysozyme produces a decrease in A350 of 0.001 per minute at 23°C with a 0.5 mg/ml suspension of lyophilized E. coli K802 cells in 50 mM Tris-HCl (pH 7.5).
Storage Buffer
50% glycerol containing 50 mM Tris-HCl (pH 7.5), 0.1 M NaCl, 0.1 mM EDTA, 1 mM DTT, and 0.1% Triton® X-100.
If you cannot find the answer to your problem then please contact us or telephone +44 (0)1954 210 200