The NxSeq™ AmpFREE Low DNA Library Kit allows you to build the best
fragment DNA libraries possible. Each step has been optimised to ensure peak performance on Illumina sequencers. In addition, these
kits require only 75 ng of sheared DNA input, and produce libraries in
about 2 hours using a streamlined, easy to follow protocol.
- Low Input: Requires as little as 75 ng of sheared input DNA allowing use of limiting samples.
- High Efficiency: Optimised adaptor ligation produces more
sequenceable fragments in each library, yielding better coverage &
depth from single or multiplexed libraries.
- PCR-free: Prevents the introduction of PCR-bias, providing more uniform coverage.
- Fast: 2 hour, 10 minute protocol saves you time and gets your samples on the sequencer sooner.
- Minimal GC bias
- Affordable: Best priced and best performing kit available.
The NxSeq AmpFREE Low DNA Library Kit is compatible with NxSeq Adaptors or other dT-tailed, Illumina-compatible adaptors.
Each NxSeq AmpFREE Low DNA Library Kit contains Enzyme Mix (EM), 2X Buffer (2XB), Ligase (LIG) and Elution Buffer (EB). Adaptors must be purchased separately.
Higher Efficiency Libraries
More Sequenceable DNA Fragments per Library = More Data
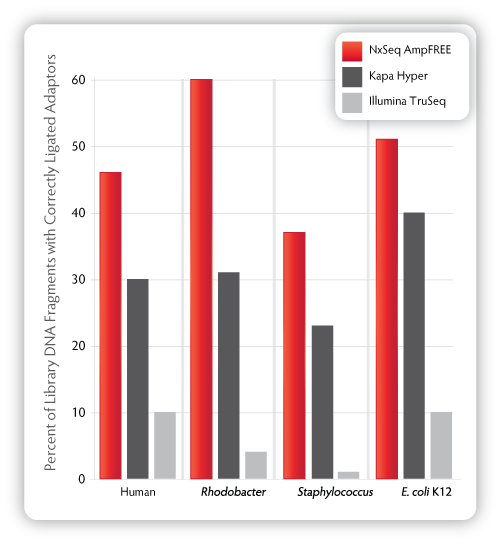
Figure 1. Percentage of library DNA with correctly ligated adaptors measured by qPCR. Duplicate libraries were prepped per kit/organism (Human, Staphylococcus aureus, Rhodobacter sphaeroides (1 library only), and E. coli)
according to the manufacturer’s recommended input amounts and
protocols. Adaptor ligation efficiency was measured by qPCR using the
KAPA Library Quantification Kit (Complete ROX Low, Item No.KK4873) and
matching amplified library as an internal standard.
Sequencing Impact of Higher Efficiency Library Construction
Better Libraries Increase the Number of Reads per Library
| Library Kit |
DNA Input |
Total Number of Sequencing Reads Per Library |
Staphylococcus aureus
|
E. coli K12
|
|
NxSeq AmpFREE Low DNA Library Kit
|
75 ng
|
5,649,946 |
4,305,882 |
| Kapa Hyper Prep Kit |
250 ng |
4,838,726 (-15%)
|
1,647,452 (-62%)
|
| Illumina TruSeq DNA PCR-Free Library Prep Kit |
1 µg |
38,768 (-99%)
|
1,543,558 (-64%)
|
Figure 2a. Number of sequencing reads generated per library after multiplexing and running on a MiSeq Instrument. DNA
fragment libraries were prepped in parallel for each kit/organism
according to the manufacturer’s recommended input amounts and protocols.
Libraries were quantitated and normalized to 2 nM using the Bioanalyzer
(size) and Qubit Fluorometer (amount). Equimolar amounts of each
library were multiplexed and sequenced with a single MiSeq run using 2
×150 bp chemistry. The number of sequencing reads obtained are shown as
well as the percent reduction (%) in total reads compared to the appropriate NxSeq AmpFREE Kit results.
More Proof with Challenging FFPE Samples
| Library Kit |
Sample Type |
Input Amount |
Total Reads |
Mapped Reads
(repeat masked) |
|
NxSeq AmpFREE Low DNA Library Kit
|
Normal gDNA |
75 ng |
2,163,636 |
900,338 |
| FFPE DNA |
75 ng |
1,767,818 |
688,074 |
| FFPE DNA |
150 ng |
1,706,714 |
656,658 |
| Kapa Hyper Prep Kit |
Normal gDNA |
250 ng |
1,567,276 (-28%)
|
650,296 (-28%)
|
| FFPE DNA |
250 ng |
1,270,870 (-28%)
|
487,872 (-29%)
|
Figure 2b. Number of sequencing reads generated from matching normal and FFPE gDNA sample libraries. DNA
fragment libraries were prepped using the two indicated kits according
to the manufacturer’s recommended input amounts and protocols. Libraries
were constructed from normal gDNA (Biochain, Cat. No. D1234142-S02) and
DNA extracted from a matching FFPE human kidney tissue (Biochain Cat.
No. T2234142-S02) using the Qiagen AllPrep DNA/RNA FFPE Kit. The gDNA
samples were sheared to ~250 bp before starting library construction.
Final libraries were quantitated and normalized to 2 nM using the
Bioanalyzer (size) and Qubit Fluorometer (amount). Equimolar amounts of
each library were multiplexed and sequenced with a single MiSeq run
using 2 × 150 bp chemistry. The number of sequencing reads obtained are
shown as well as the percent reduction (%) in total and mapped reads compared to the corresponding NxSeq AmpFREE Kit results using 75 ng of input DNA.
Highly Mappable Reads (>92%) from Human, Staphylococcus and Rhodobacter gDNA Sequencing
|
Sequencing Stat
|
Human |
Staphylococcus
|
Rhodobacter
|
|
Genome size, GC percentage
|
~3 Gbp 45% GC
|
2,821,361 33% GC
|
4,602,977 69% GC
|
|
Raw reads
|
3,131,114
|
1,260,836
|
3,900,174
|
|
Mapped reads
|
2,979,237 (95.15%)
|
1,174,111 (93.12%)
|
3,613,165 (92.64%)
|
|
Read length
|
148.9 bp
|
148.8 bp
|
149.6 bp
|
|
Total bases
|
443,767,447
|
174,694,261
|
540,403,552
|
|
Genome fraction
|
0.11
|
0.97
|
1.00
|
|
Avg. coverage
|
0.15X
|
62X
|
117X
|
Figure 3. Representative gDNA sequencing stats from three different organisms. Genomic
DNA fragment libraries were generated using the NxSeq AmpFREE Low DNA
Library Kit using 75 ng of sheared gDNA input from three organisms
(human, Staphylococcus aureus, and Rhodobacter sphaeroides).
The final libraries were quantitated and normalized to 2 nM final
concentrations using the Bioanalyzer and Qubit fluorometer, and 5 µL of
each library was run on a MiSeq using 2 x 150 bp chemistry and analysed.
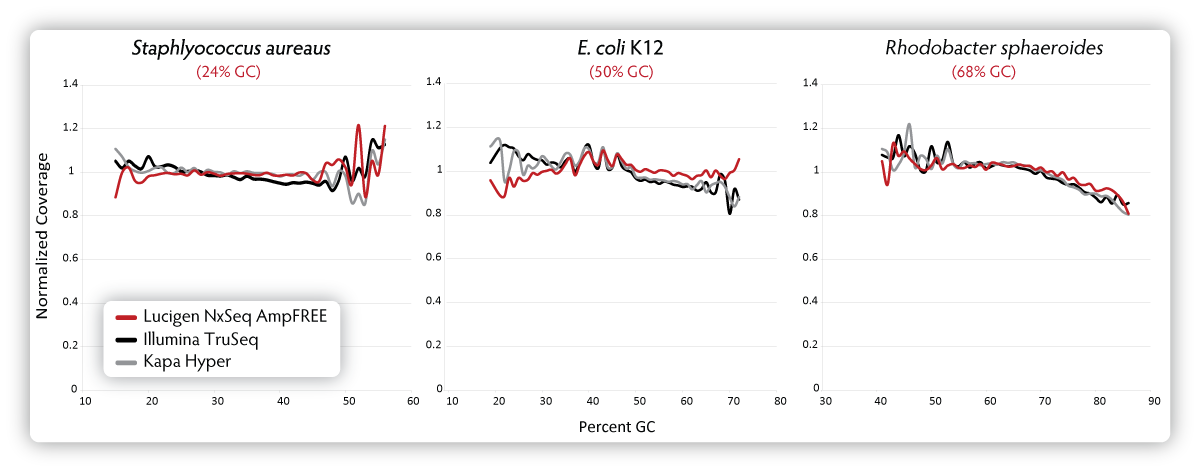
Figure 4. Sequencing bias measured for three different organisms with varying GC content. DNA fragment libraries were generated from gDNA of three organisms with varying GC content ( Staphylococcus aureus, 24% GC; E. coli K12, 50% GC; and Rhodobacter sphaeroides,
68% GC) according to the manufacturer’s recommended input amounts and
protocols. Samples were quantitated using the Bioanalyzer and Qubit
fluorometer and normalized to 2 nM final concentrations. Five µL of each
library sample was sequenced on a MiSeq using 2 x 150 bp v2 chemistry
and analysed. Normalized coverage was calculated as the (average
coverage of all windows with X% GC content) divided by the (overall
average coverage).
Faster Protocol with Significantly Less Hands-on Time
Save Time and Get Your Samples on the Sequencer Sooner!