OmniCleave™ Endonuclease is a highly purified enzyme from a recombinant E. coli strain that degrades single- and double-stranded DNA and RNA to di-, tri-, and tetranucleotides(Figure 1). OmniCleave™ Endonuclease has the same substrate specificity and yields the same products as benzonase®, an enzyme derived from Serratia marcescens.
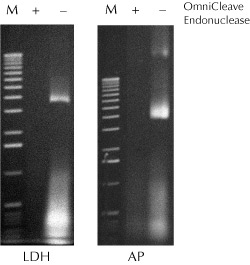 |
Figure 1. Removal of nucleic acids from cell lysates using OmniCleave™ Endonuclease. Cell lysates were prepared with or without OmniCleave Endonuclease from E. coli cells expressing human lactate dehydrogenase B (LDH) and E. coli alkaline phosphatase (AP). Two microliters of each of the cell lysates were separated by electrophoresis on a 1% agarose gel and the nucleic acids detected by ethidium bromide staining. Lane M, 1-kb ladder |
Unit Definition:
One unit converts 1.0 OD260 (~60µg) of salmon sperm DNA into acid-soluble nucleotides in 30 minutes at 37°C in 50mM Tris-HCl, pH 8.0 at 25°C and 1mM MgCl2.
Storage Buffer:
50% glycerol solution containing 50mM Tris-HCl, pH 7.5 at 25°C, 0.1M NaCl, 0.1mM EDTA, 1mM dithiothreitol, and 0.1% Triton® X-100.
Quality Control:
OmniCleave™ Endonuclease is free of detectable protease activity.
If you cannot find the answer to your problem then please contact us or telephone +44 (0)1954 210 200