.
EZ-QC™ mRNA Capping Efficiency Assay Kit
- Quantitate mRNA capping efficiency (percent capped RNA content) using standard polyacrylamide gel electrophoresis (PAGE).
- Easy, cost-effective, benchtop method useful for QC as an alternative to HPLC and LC-MS analysis.
Product Description
The EZ-QC™ mRNA Capping Efficiency Assay Kit provides quantitative
capping efficiency (percent
capped RNA content) analysis of synthesized
mRNA. 5'-cap structures can be added co-transcriptionallyor
post-tran
scriptionally to in vitro transcribed (IVT) RNA to
produce Cap 0 and/or Cap 1 structures as a step in the mRNA synthesis
process. Since only capped RNAs will be expressed in cells, it is
important to have the highest possible percentage of capped RNAs present
in a sample. The EZ-QC™ mRNA Capping Efficiency Assay Kit simplifies
analysis and replaces tedious and expensive capping efficiency
determination methods such as HPLC and mass spectrometry, instead
allowing for determination based upon standard polyacrylamide gel
electrophoresis (PAGE) methodology.
The EZ-QC™ mRNA Capping Efficiency Assay utilizes a chimeric
RNA:DNA:RNA Targeting Oligonucleotide (user provided) which hybridizes
to the 5'-end of an RNA mixture. This hybridization complex serves as a
substrate for RNase H cleavage releasing the capped and uncapped 5'-end
fragments which can subsequently be resolved via PAGE and quantified by
gel band analysis allowing for the calculation of the percent capped RNA
content of the sample. The EZ-QC™ mRNA Capping Efficiency Assay does
not differentiate between Cap 0 and Cap 1 capped RNAs, only between
capped (Cap 0 Cap 1) and uncapped RNAs.
For determination of Cap 0 vs Cap 1 content, CELLSCRIPT™ also offers the EZ-QC™ mRNA Cap 1 Efficiency Assay Kit as well as the EZ-QC™ mRNA Poly(A) Tail Length Assay Kit for complete mRNA characterization.
Product Performance
EZ-QC™ mRNA Targeting Oligonucleotide design considerations
The EZ-QC™ mRNA Capping Efficiency Targeting Oligonucleotide should
be designed to release an mRNA 5'-end fragment of 25-40 nucleotides in
length (Figure 1). This released fragment, /– the cap nucleotide, is to
be resolved via PAGE analysis. RNase H cleavage will occur in the mRNA
strand across from the 5'-most DNA base in the Targeting
Oligonucleotide.
Figure 1. Graphic representation of mRNA and Targeting Oligonucleotide.

Resolution of capped and uncapped 5’-end fragments
Mixtures of capped and uncapped mRNA were assayed using the EZ-QC™
mRNA Capping Efficiency Assay Kit and resolved on a polyacrylamide gel
(Figure 2A). The Targeting Oligonucleotide facilitates cutting the mRNA
at a single, repeatable location allowing for greater confidence in
determining the percentage of capped and uncapped content of the assayed
samples. The percent capped and uncapped mRNA in a mixture are
calculated with a simple readout from any gel imaging system (Figure
2B).
Figure 2A. Capped
and uncapped 5’-end fragments are efficiently resolved by PAGE allowing
for capping efficiency analysis with standard lab equipment.
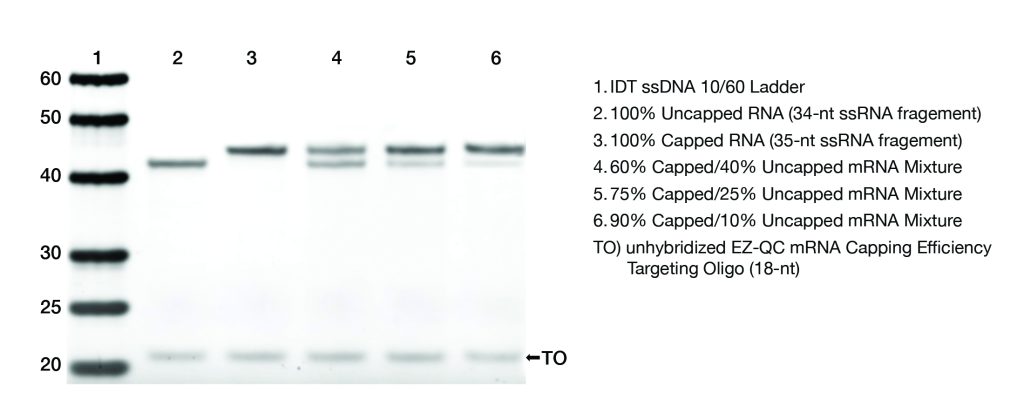
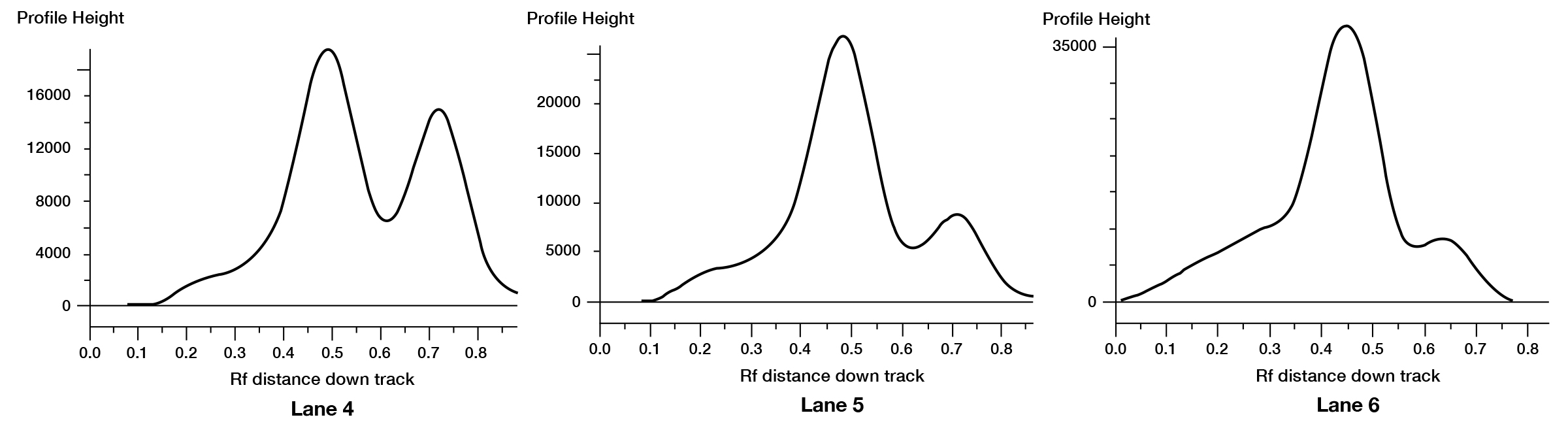
Figure 2B. Densitometric data from lanes 4, 5, and 6 from the above PAGE gel using a Syngene® G:Box Gel Documentation System. The percent capped and uncapped mRNA in each mixture are calculated using a simple readout.
Materials Supplied
Important Store at -20°C in a freezer without a defrost cycle. Do not store at –70°C.
EZ-QC™ mRNA Capping Efficiency Assay Kit (10 reactions)
Sufficient for 10 experimental and 10 control reactions. |
|
Kit Component |
Reagent Volume |
EZ-QC™ RNase H
in 50%
glycerol, 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM dithiothreitol
(DTT), 0.1 mM EDTA and 0.1% Triton® X-100. |
20 μl |
10X EZ-QC™ RNase H Reaction Buffer
0.2 M Tris-acetate, pH 7.9, 0.5 M potassium acetate, 0.1 M magnesium acetate and 0.01 M DTT. |
20 μl |
ScriptGuard™ RNase Inhibitor, 40 U/μl
in 50% glycerol, 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM DTT, 0.1 mM EDTA and 0.1% Triton X-100. |
10 μl |
Stop/Loading Buffer
95% formamide, 10 mM EDTA, pH 7.5, 0.01% Bromophenol Blue and 0.01% Xylene Cyanol |
200 μl |
|
RNase-Free Water |
500 μl |
Materials Required, but not Supplied
- Purified capped mRNA/RNA
- EZ-QC™ mRNA Capping Efficiency Targeting Oligonucleotide
- Materials for polyacrylamide gel electrophoresis.
- Materials for polyacrylamide gel visualization, imaging and quantification.
- Optional: molecular weight marker (e.g., ssDNA 10/60 Ladder [IDT])
Downloads
Manual
SDS
FAQs: For answers to Frequently Asked Questions about our products, visit the FAQ library.
If you cannot find the answer to your problem then please contact us or telephone +44 (0)1954 210 200