Plasmid-Safe™ ATP-Dependent DNase selectively removes contaminating bacterial chromosomal DNA from plasmid, cosmid, fosmid, and BAC clones or vector preparations. Such preparations are frequently contaminated with fragments of bacterial genomic DNA generated during alkaline lysis. Other purification options, such as spin-columns or even CsCl centrifugation, do not effectively remove these contaminants and require further purification steps. Contaminating DNA fragments left behind by these methods ultimately can become ligated into a cloning vector, resulting in false positives and high backgrounds, or erroneous sequence data.
Plasmid-Safe ATP-Dependent DNase digests linear dsDNA to deoxynucleotides at slightly alkaline pH and, with lower efficiency, closed-circular and linear ssDNA. The enzyme has no activity on nicked or closed-circular dsDNA or supercoiled DNA. Therefore, Plasmid-Safe DNase is ideal as the final purification step for plasmid, cosmid, fosmid, and BAC vector and clone preparations
| Figure 1. Plasmid-Safe™ ATP-Dependent DNase removes contaminating genomic DNA from plasmid preps. Lane 1, 3 µg of Sma I-digested bacterial chromosomal DNA; lane 2, 500 ng of uncut plasmid DNA; lane 3, mixture of 3 µg of digested bacterial chromosomal DNA and 500 ng of uncut plasmid before Plasmid-Safe DNase treatment; lane 4, mixture of chromosomal DNA and plasmid DNA after Plasmid-Safe DNase treatment (incubation with Plasmid-Safe DNase for 30 minutes at 37°C); lane M, Kilobase ladder |
Unit Definition
One unit converts 1 nmole of deoxynucleotides in linear T7 DNA into an acid-soluble form in 30 minutes at 37°C using the prescribed assay conditions. Three units will digest 1µg of DNA in 30 minutes at 37°C.
Storage Buffer
50% glycerol containing 50mM Tris-HCl, pH 7.5, 0.1 M NaCl, 0.1mM EDTA, 1mM DTT and 0.1% Triton® X-100.
Plasmid-Safe 10X Reaction Buffer
330 mM Tris-acetate, pH 7.8, 660mM potassium acetate, 100mM magnesium acetate, and 5.0mM DTT. ATP must be added to a final concentration of 1mM in the 1X Buffer.
Quality Control
Plasmid-Safe DNase is free of detectable RNase and double-stranded, DNA-specific endonuclease activities.
Plasmid-Safe Protocol Outline
- Isolate DNA from bacteria using standard mini- or maxi-prep protocols
- Resuspend the pelleted DNA in 1X Plasmid-Safe Reaction Buffer with 1mM ATP.
- Add Plasmid-Safe DNase.
- Incubate at 37°C: 15 minutes for mini-prep DNA or 2 hours for DNA from a 500-ml prep.
- Inactivate Plasmid-Safe DNase by incubation at 70°C for 30 minutes.
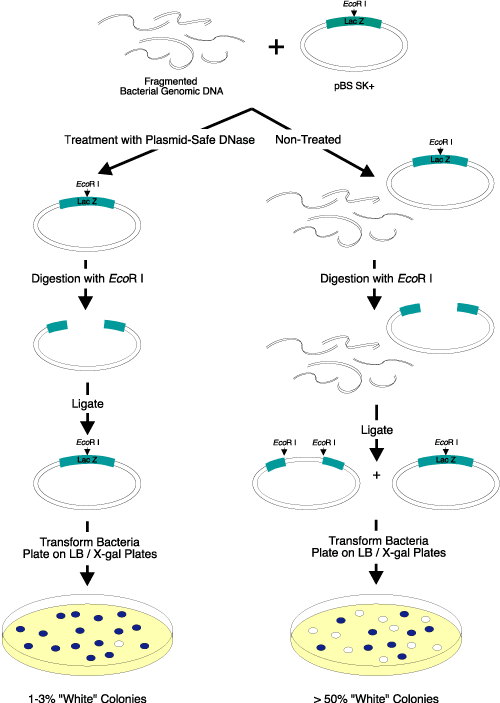 |
Figure 2. Elimination of linear DNA resulting in white colonies. Three micrograms of EcoR I-digested bacterial genomic DNA were added to
2 µg of a supercoiled lacZ-containing plasmid vector. Half of the DNA mixture was treated with Plasmid-Safe DNase; the other half was not treated and served as the control. After heat inactivation of Plasmid-Safe DNase, the DNA was digested with EcoR I, ligated overnight with T4 DNA Ligase (EPICENTRE), and transformed into competent cells. The transformants were plated on IPTG/X-gal-containing medium. Only 1-3% of the colonies transformed by the Plasmid-Safe DNase-treated DNA were white, while greater than 50% of the colonies transformed by the control DNA sample (untreated) were white. Use of Plasmid-Safe DNase resulted in elimination of almost
all of the linear DNA. |
If you cannot find the answer to your problem then please contact us or telephone +44 (0)1954 210 200